Research Article - (2023) Volume 5, Issue 3
Dental caries is now one of the world's most common chronic diseases. Caries are irreversible diseases caused by external factors that do not manifest symptoms at first. In terms of detailed examination of etiological factors and taking necessary precautions, great strides have been made in preventive dentistry over the years. However, another point to investigate is the hereditary aspect of dental caries and what kind of genetic changes it causes. Tooth decay, trauma and other factors can all harm pulp tissue. Infection in the pulp tissue may cause tissue degradation via molecular and/or cellular events, causing the dental pulp's inflammatory response to be regulated by genetic and epigenetic events.
Deep dentine caries • Epigenetic modifications • Acetylation• Enflammation 8 • Cytokines
Epigenetics is the study of changes in gene expression that are inherited but are not caused by changes in the DNA sequence. Molecular mechanisms behind epigenetic processes are complicated and involve histone modifications generated by noncoding RNAs, DNA methylation, histone variant placements and gene regulation amongst other mechanisms. Epigenetic changes have a significant impact on gene expression. Infections and other environmental factors are also known to have an impact on epigenetic changes and diseases [1].
The effects of histone modifications on gene expression vary. Histone methylation on histone H3 has been linked to dental pulp inflammation and repair processes. Tri-methylation of lysine 27 on histone H3 protein (H3K27me3) levels have been found to be lower in inflamed pulp tissue and pulp cells, which can trigger the process of dental pulp tissue regeneration [2].
Histone acetylation is another important histone modification. Two groups of cellular enzymes, Histone Acetyltransferases (HATs) and Histone Deacetylases (HDACs), balance the acetylation process. Histone Deacetylase inhibitors (HDACis) have been presented as a tool of epigenetically inducing reparative processes in dental pulp cells by decreasing proliferation and enhancing mineralization in these cells. CREB-Binding Protein (CBP/p300), a HAT known to inducetranscriptional activation, rapidly acetylates the promoters of various pro-inflammatory cytokines (IL-12, IL-8, IL-2, and IL-1) [3].
Recent evidence suggests that histone acetlytransferase HAT may play a significant role in pulp inflammatory processes, explaining the importance of further findings into these mechanisms. Following inflammation, acetylation of histone H3 at the promoters of several cytokines and chemokines may result in increased Nuclear Factor Kappa B (NFk-B) to these sites. According to the findings, histone acetylation modulation can be used in clinical practice in restorative dentistry for inflammation control, repair and regeneration. Furthermore, these inflammatory diseases are characterized by local accumulation of inflammatory mediators such as cytokines and chemokines, which actively participate in the destructive and reparative processes in the pulp [4].
Cytokines are small proteins that are secreted by cells and have a specific effect on cell interactions and communication. Pro-inflammatory cytokines are primarily produced by activated macrophages and play a role in enhancing inflammatory responses. There is a lot of evidence that proinflammatory cytokines like IL-1, IL-6 and TNF-alfa are involved in the pathological pain process. Interleukin 6 (IL-6) is a pro-inflammatory cytokine that also functions as an anti-inflammatory myokine [5].
Many studies have found an increase in Tall Like Receptor (TLR) expression in inflammatory pulp tissues. TLRs are involved in osteogenesis and odontogenesis, both of which are important in pulp tissue repair. In macrophages and endothelial cells, Interferon gamma (IFN-gamma) acts as an inflammatory mediator in the initial pulpal response to caries. IFN-gamma may have an effect on the immune response during pulpitis [6].
The presence of an inflammatory reaction in the dental pulp was linked to a loss of DNA methylation in the IFN-gamma promoter region. The transition from a fully methylated to a partially methylated IFN-gamma state characterizes inflammation in dental pulp tissue. These findings suggested that epigenetic events in the dental pulp may be related to IFN-gamma modulation [7].
The aim of this study is to compare the genetic changes in pulp tissue from healthy teeth and pulp tissue from teeth with deep dentin caries, based on histone acetylation and the presence of cytokines whose secretion has been proven in the inflammatory response. Another goal is to learn more about the epigenetic mechanisms that cause inflammation in pulp tissue by examining the histone acetylation pattern in healthy pulp tissue and pulp tissue with deep dentin caries. The null hypothesis of this study is that histone H3 gene expression, inflammatory cytokines (IL-6, TLR2, IFN-gamma), mRNA expression and protein secretion differ between healthy pulp tissue and pulp tissue samples with deep dentin caries [8].
Tissue collection, preperation and isolation
The ethics committee decision, numbered 2019/611 and dated 11.09.2019, approved this study. The research was conducted at faculty of dentistry and faculty of medicine's department of physiology [9].
Tissue samples from 20 different patients were collected from the Healthy Pulp (HP) group. Patients' samples were obtained from impacted and semi-impacted third molars and extractions were recommended for patients with impacted teeth according to the position of the existing teeth in the jaw and the maxillary and mandibular arches were not differentiated. Ten HP tissue samples were used for RT-qPCR and ten were used for western blot analysis [10].
Affected Pulp (AP) tissue samples from 20 different patients' teeth with deep dentin caries were obtained. The patients' samples were taken from their third molar teeth, with an indication for extraction based on their clinical findings. There was no distinction between the maxillary and mandibular arches. Ten AP tissue samples were used for RT-qPCR and ten were used for Western blot analysis [11].
Just after extraction, the teeth were cleaned with 70% ethyl alcohol and soft tissue residues were removed. Small incisions were made 5 mm apically from the enamel-cementum junction with diamond burs with the use of water cooling (without damaging the pulp). Pulp tissues were obtained after the crown of the tooth was broken through the incision line with surgical forceps. Tissue samples were stored at 80°C after being kept at 2°C -8°C overnight [12].
Real time quantitative RT-PCR
Total RNA isolation from dental pulpal tissue: Frozen pulp tissues (ten with deep dentin caries and ten with healthy dental pulp), each in its own purezol tube for RNA isolation. Following that, the tissues were homogenized using an injector. After adding 200 l of chloroform, the mixture was vortexed for 15 seconds. It was incubated at room temperature for 15 minutes. It was centrifuged for 20 minutes at +4°C at 12000 g. Afterward, the aqueous phase was transferred to a larger 1.5 mL tube and about 300 l of isopropanol (depending as to how much water there was) was added to the new tube. Then the tube was turned upside down a few times. After that, the tubes were centrifuged at 12000 g for 10 minutes at +4°C. The supernatant that was left over at the end of the centrifugation process was thrown away. 1 ml of 75% ethanol was lightly stirred into the pellets at the bottom. The pellet was then washed by centrifuging it at 7500 g for 5 minutes at +4°C. After centrifugation, the supernatant was discarded and the ethanol surrounding the pellet was pipetted and cleaned. For 5 minutes, the tubes were dried at room temperature until the ethanol evaporated. RNA-containing pellets were dried and after that 20 l of NFW (Nuclease-Free Water) was added to them and the pellet was mashed up again. Purities and concentrations of RNA (ng/l) isolated with nanodrop were measured while the samples were on ice [13].
cDNA synthesis
Following the iScript cDNA synthesis kit protocol, cDNA was synthesized from RNA samples at specified concentrations. The cDNA synthesis cocktail, the contents of which are listed in Table 1, was made on ice without the inclusion of template RNA. Each labeled PCR tube received 15 L of the prepared cocktail. Each RNA sample received 5 L (1000 ng) in its own tube. The CFX connect real-time PCR detection system (BioRad) was usption on the parameters shown in Table 2.
| cDNA cocktail content | Volume (µL) |
|---|---|
| Reaction buffer (5x) | 4 |
| Template RNA | 5 |
| Water, PCR | 9 |
| Enzyme mix (10x) | 2 |
| Total | 20 |
Table 1: cDNA synthesis cocktail.
| Step | Temperature | Time (minute) |
|---|---|---|
| 1 | +42°C | 15 |
| 2 | +85°C | 5 |
| 3 | +65°C | 15 |
| 4 | +4°C | Indefinite |
Table 2: Reverse transcription protocol.
RT-qPCR
Quantitative RT-PCR studies were performed on the CFX connect real-time PCR detection System device, using the SsOAdvanced Universal IT SYBR Green Supermix (BioRad) kit, to examine changes in the expression of target genes. The TATA Box Binding Protein (TBP) reference gene, the primer sequence for which is shown in Table 3, was used as the control gene. Ct (threshold cycle) values were calculated using the 2-ΔΔCT method and normalized with reference gene Ct values at the end of the procedure. The target genes were histone H3-4, IL-6 and TLR2. Table 4 lists the primer sequences in detail.
| Reference gene | Primer position | Primer sequence |
|---|---|---|
| TBP | GAGTTCTGGAAGGTTCAGGTTG | 5'…3' |
| CGTGGTTTCGTGGCTCTCTT |
Table 3: Primer sequences for reference gene.
| Target genes | Primer | Primer sequence |
|---|---|---|
| H3-4 | CAAGGTGGCTCGAAGA | 5'..3' |
| GCGCAGGTCGGTCTTAAA | 5'..3' | |
| IL6 | ACTCACCTCTTCAGAACGAATTG | 5'..3' |
| CCATCTTTGGAAGGTTCAGGTTG | 5'..3' | |
| TLR2 | GGAAGCTGGTGGCAATAACT | 5'..3' |
| GTGCTGTCCTGTGACATTCC | 5'..3' |
Table 4: Primer sequences for target genes.
Western blot analysis
Protein isolation from dental pulpal tissues was carried out using the RIPA buffer. The tissues were lysed in RIPA buffer, which was bolstered with a protease inhibitor cocktail, PMSF and sodium orthovanadate by adding 10 ml of each to 1 ml of RIPA on ice immediately before use and homogenized by mechanical homogenization after being located on ice for 10 minutes. The homogenates were centrifuged at 15,000 × g for 20 min at 4°C. Bicinchoninic Acid (BCA) reagent was used to determine the concentrations of isolated proteins. SDS-PAGE gels were loaded with 50 g of protein per well. Proteins were separated electrophoretically using a Biorad electrophoresis system set to 100 maximum volts and 25 mA per gel. The separated proteins were transferred from the gel to the PVDF membrane after electrophoresis; blot paper, PVDF membrane, gel and blot paper were transferred by sequencing. TBS (20 mM Tris, 0.5 M NaCl) containing 5% (w/v) skimmed milk powder was used to block the membrane for 1 hour. It was then washed five times for 25 minutes in 5 min. interval with TBS (TBS-T) containing 0.5 percent (v/v) Tween 20 (Sigma, P1379). The membrane was incubated with primary antibody at +4°C for 12-16 hours (overnight incubation) (Table 5).
| Target antibody | Primer antibody | Dilution ratio |
|---|---|---|
| Acetyl H3 | Acetyl-histone H3 (Cell signaling, Lys9/Lys14) antibody | 1:2000 |
| IL-6 | Anti-IL-6 (Santa Cruz, sc-130326) antibody | 1:2000 |
| TLR2 | Anti-TLR2 (Santa Cruz, sc-21759) antibody | 1:2000 |
| IFN-gamma | Anti-IFN-γ/Interferon gamma (Santa Cruz, sc-8423) antibody | 1:2000 |
Table 5: Target antibody information.
The reference protein was beta-actin (β-actin) antibody diluted (1/500) in TBS-T. Blotting was done visualized using the SynGeneG Box system, which is available at Erciyes university's faculty of medicine, department of physiology. The optical density of certain bands in the membrane was determined using Image J software (National Institutes of Health, USA), which was then normalized to the optical density of beta-actin and reported as an arbitrary unit relative to a sample of control group in each membrane.
Statistical analysis
The data's conformity to the normal distribution was analyzed using a histogram, q-q graphs and the Shapiro-Wilk test. Levene's analysis was used to measure variance homogeneity. The independent t-test was used to compare statistically between groups (2 group levels) in tissue samples from Healthy Pulp (HP) and Affected Pulp (AP). GraphPad Prism software was used to analyze the data (San Diego, CA). P<0.05 was considered as statistically significant.
RT-qPCR analysis results
When we look at results of the RT-qPCR analysis, we see that while IL-6 gene expression was higher in the AP group than in the HP group, the difference was not statistically significant (p>0.05) (Figure 1).
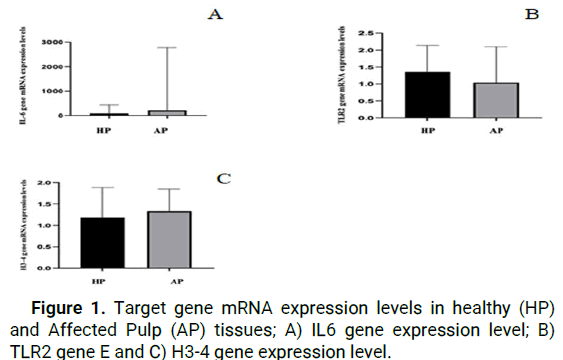
Figure 1: Target gene mRNA expression levels in healthy (HP) and Affected Pulp (AP) tissues; A) IL6 gene expression level; B) TLR2 gene E and C) H3-4 gene expression level.
Fold change analyses indicated relative mRNA expression changes between groups. The IL-6 gene expression level was 3.40 in HP and 26.57 in EP, demonstrating increased expression in both target tissues.
Following an investigation of the levels of mRNA expression of the TLR2 gene, it was discovered that the mRNA expression levels in the samples belonging to the HP group were considerably higher than those belonging to the AP group. When the findings were analyzed by using fold change method, it was discovered that the expression of the TLR2 gene was 1.31 positive values in the HP group and 1.21 positive values in the AP group. According to the results of the statistical analysis, there was no statistically significant difference between the HP and AP groups (p>0.05) (Figure 2).
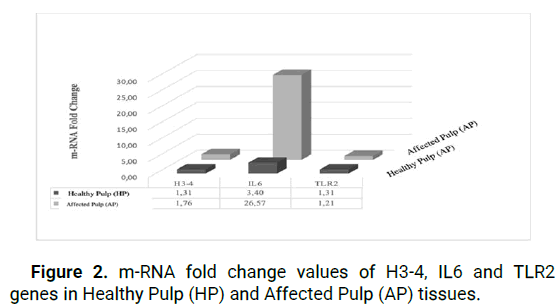
Figure 2: m-RNA fold change values of H3-4, IL6 and TLR2 genes in Healthy Pulp (HP) and Affected Pulp (AP) tissues.
As a result of the RT-qPCR analysis, it was reported that Histone H3-4 gene expression was higher in AP tissue cells as compared to HP tissue cells at the mRNA expression level of the histone H3-4 gene. The use of fold change analysis provided additional evidence for the existence of relative changes in mRNA expression levels. The level of H3-4 gene expression was determined to be 1.31 positive values in the HP group and 1.61 positive values in the AP group, with a significant rise in the expression of the H3-4 gene recorded in both target tissues (p>0.05).
Western blot analysis results
When the results of our research were analyzed, IL-6 protein expression was shown to be higher in cells from AP tissue as compared to cells from HP tissue, however this difference was not statistically significant (p>0.05) according to Independent t-test (Figure 3).
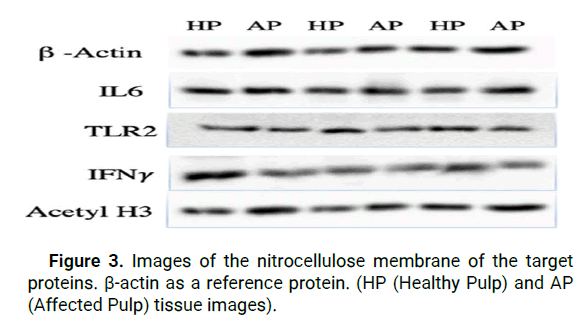
Figure 3: Images of the nitrocellulose membrane of the target proteins. β-actin as a reference protein. (HP (Healthy Pulp) and AP (Affected Pulp) tissue images).
As an outcome of the analysis, it was discovered that the expression of TLR2 protein was higher in cells from HP tissue as compared to cells from AP tissue in western blot analyses, but that this difference was not statistically significant (p>0.05).
The results of Western blot analysis of TLR2 cytokine and mRNA expression levels obtained from RT-qPCR analysis are consistent and support the data obtained.
Further evaluation of our study's findings indicated that IFN-δ protein expression was surprisingly high in HP tissue cells when compared to AP tissue cells, however this difference was not statistically significant (p>0.05).
The protein expression represented by the acetylated histone H3 gene was found to be significantly higher in the AP group as compared to the HP group (p=0.01), indicating that the acetylated histone H3 gene is more active (Figure 4).
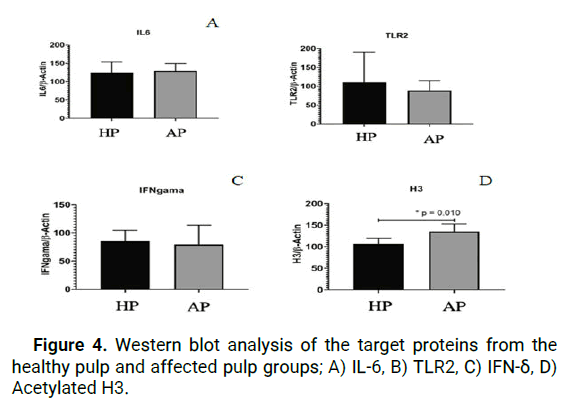
Figure 4: Western blot analysis of the target proteins from the healthy pulp and affected pulp groups; A) IL-6, B) TLR2, C) IFN-δ, D) Acetylated H3.
The aim of this study was to compare the acetylation patterns of histone H3 (acetylated) gene in healthy dental pulp tissue cells and deep dentin caries (D3) pulp tissue cells. Additionally, we investigated the gene and protein secretion of cytokines and surface receptors such as IL-6, IFN-δ and TLR-2. In Western blot analysis, our hypothesis was supported since the acetylation of the H3 gene protein has been shown to be statistically significant in the deep dentin caries group, yet our hypothesis was not supported because there was no statistically significant difference between the groups in other analyses.
In order to better understand the inflammation-associated repair mechanisms of pulpitis cellular regulators and develop new therapeutic options, prospective and biological inductionbased research is required. In recent years, new technologies and research areas have revealed exciting discoveries about new dental biomaterials that are effective in the pulp tissue regeneration process, potentially eliminating the need for root canal treatment for patients who are experiencing clinically symptomatic dental caries. The effects of epigenetic modifications to DNA-associated proteins and non-coding RNAs have been shown to regulate mineralized tissue development and repair, as well as control pulp tissue cell and stem cell populations.
Studies focusing on the DNA-related histone acetylation process, which is stabilized by Histone Acetyltransferases (HAT) and Histone-Deacetylases (HDAC), have emerged as a potential new solution. Recent research suggests that changing this balance promotes dental pulp cell mineralization and repair processes in damaged and/or caries-affected pulp. The histone H3 gene encodes a protein that is thought to be involved in the dynamic and long-term regulation of genes. Irregularities in histone acetyltransferase and histone deacetyltransferase enzyme activity play an important role in pulp tissue diseases and cell inflammatory responses. In our study, we looked at the mRNA expression of the histone H3-4 gene in healthy and affected pulp (deep dentinal caries) tissue cells using RT-qPCR, as well as the acetylation of H3 protein using western blot.
In RT-qPCR and fold change analysis, mRNA expression were found to be higher in the AP group than in the HP group. Furthermore, acetylation of H3 gene protein, was found to be significantly higher in the AP group than in the HP group. Gopinathan, et al., reported findings in dental pulp cell studies that histone acetylation and methylation are involved in dentin development and repair. Furthermore, H3K27me3 (a marker indicating tri-methylation of lysine 27 on histone H3 protein; a marker of inactive gene transcription) suppresses dentin sialophosphoprotein and dentin matrix acidic phosphoprotein-1, which are mineralization markers associated with odontoblast activity, in dental follicle stem cells.
They hypothesized that it could be an indicator of the cell repair process continuing. Gu, et al., reported that in the study groups where the acetylation mechanism was induced in the western blot findings of DPKH, acetyl H3 protein secretion was higher than in the groups where acetylation was suppressed. In light of the above findings of our own and other studies, the fact that acetylated histone H3 protein secretion was higher in the cells of the affected pulp tissue than in the cells of the healthy pulp tissue suggested that the cells of the affected pulp tissue may be more active in terms of inflammatory reactions. Again, the fact that mRNA expression levels in the H3-4 gene were higher in the affected pulp cells compared to the healthy pulp group provided support for the supporting data that gene transcription and repair mechanisms are active in the presence of caries.
Cytokines play an important role in the formation of the immune response against pathogenic agents and have the potential to control the intensity and duration of the inflammatory response. As a result, in our study, we needed to look at the levels of IL-6 cytokines, which play important roles in the inflammatory process. According to our results, the affected pulp tissue group had greater expression and protein secretion of IL-6 cytokinin than the healthy pulp tissue group, as shown by our RT-pcr and Western blot analyses. ElSalhy, et al., studied the levels of cytokines in blood tissue isolated from the pulp of healthy teeth with caries exposure and irreversible pulpitis. The levels of IL-6 in the blood of carious exposed pulp tissue were found to be 20 times higher than in healthy pulp tissue. They proposed that the findings of their study could improve the long-term prognosis of direct pulp therapy. According to this study, we found that the relative increase in IL-6 cytokine shows that findings may change in the inflammation response according on caries depth, as demonstrated by our study results. IFN-δ is involved in the pathogenesis of periapical lesions that affect dental pulp tissue and surrounding tissues. Furthermore, the IFN-δ gene's methylation and deacetylation have been linked to transcriptional inactivation. Western blot analysis revealed that IFN-δ protein expression was lower in the affected pulp group compared to the healthy pulp group, but this difference was not statistically significant between the groups in our study. Campos compared the pulp tissues of teeth with periapical lesions and teeth with perapical granulomas. The mRNA expression level of the IFN-δg gene did not differ significantly between the two study groups, according to the researchers. Cardoso examined the DNA methylation of the IFN-δ gene in the pulp tissues of healthy and irreversible teeth with pulpitis; the results show that there was no significant difference. Based on the literature and our research results, we presumed that in the presence of advanced caries lesion, IFN-δ tends to decrease over time by order to fulfill the function of cytokine and chemokine stimulation, which occurs as a result of the response to stimuli at the cellular level. Molecular patterns associated with pathogens are detected by Toll-Like Receptors (TLRs), which are members of the interleukin-1 transmembrane receptor family of transmembrane receptors. TLR-2 is primarily involved in the activation of immune responses against gram-positive bacteria and surface receptors such as TLR-2 and TLR-4 are involved in the activation of immune cells by microorganisms in the dental pulp. TLR2 surface receptor was included in our study for these reasons in order to evaluate the inflammatory response that may develop at different levels among our study groups. As a result of our study, RTqPCR and Western blot data revealed that TLR2 cytokine expression was lower in the AP group compared to the HP group, but the difference was not statistically significant. Cardoso used RT-qPCR to examine methylated (inactive gene transcription) and unmethylated (active gene transcription) TLR2 and TLR4 surface receptors on healthy and irreversible pulpitis cells. The researchers looked into the differences in DNA frequencies. They stated that the data for unmethylated DNA frequency was similar between the groups. However, in terms of both TLR-2 and TLR-4 (CD-14) surface receptors, DNA methylation patterns did not differ statistically between healthy and pulpitis tissue samples. Mutoh used RT-PCR and immunohistochemical analyses to investigate the mRNA expression of TLR-2 and TLR-4 in carious pulp cells after creating an experimental pulp perforation in BALB-c mice an d a dental caries model with exposed cavity preparation. TLR-2 and TLR-4 were expressed in the early stages of pulpitis in these studies and TLR2 mRNA expression in pulp tissue of teeth with pulpitis began to increase 3 hours after infection, reached a peak at 9 hours, gradually decreased after 9 hours and decreased at 72 hours. It has been reported that there is a significant tendency for expression levels to decrease. When these studies in the literature and our data are compared, it is hypothesized that the TLR-2 surface receptor will show a decreasing trend after completing the pathogen recognition task in the tissue and will not increase retrospectively with the completion of the bacterial infiltration process.
To sum up our findings, the IL6 and H3-4 genes, active expression was seen in damaged pulp tissue, whereas TLR2 was prominent in healthy pulp tissue. It was shown that the affected pulp tissue had increased levels of IL-6 and acetylated H3 protein, in contrary TLR and IFN-γ proteins released more in healthy pulp tissue. Examining the study groups with different dentin caries levels and reversible-irreversible pulpitis with similar methodology in prospective studies suggested that the data that was felt to be lacking in the research could be obtained in our study.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Melike D, et al. "Acetylation Pattern and Cytokine Secretion in Teeth with Deep Dentine Caries". J Dent Res Pract, 2023, 5(3), 1-5.
Received: 12-Jun-2023, Manuscript No. JDRP-23-102214; Editor assigned: 15-Jun-2023, Pre QC No. JDRP-23-102214 (PQ); Reviewed: 29-Jun-2023, QC No. JDRP-23-102214; Revised: 13-Sep-2023, Manuscript No. JDRP-23-102214 (R); Published: 11-Oct-2023, DOI: 10.4172/jdrp.23.5 (3).046
Copyright: © 2023 Melike D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.