Review Article - (2021) Volume 10, Issue 3
Poly (ADP-Ribose) polymerase • Ischemia-reperfusion • Infarction • Diabetes • Hypertension • Inflammation • PARP inhibitors • Necrosis
Chambon and his colleagues invented the addition of NAD+ to rat liver nuclear extracts, stimulated the synthesis of a polyadenylic acid. This polyadenylic acid was later recognized as Poly(ADP-Ribose) or PAR [1]. This invention led to the increased interest of researchers in the area of Poly(ADP-Ribosyl)ation and led to the invention of the first PAR Polymerase (PARP) isoform which is now termed as PARP1. Discovery of PARP1 led to further discovery of several other PARP family members. Poly (ADP-Ribose) Polymerase (PARP) are the proteins which are involved in various physiological and pathophysiological pathways including DNA damage signaling, cell death pathways, for the maintenance of cell proliferation, differentiation, neuronal function and inflammation. Further approaches in this area led to the identification of 18 putative PARP sequences in the human genome. Good amount of information is available for the first 5 enzymes i.e., PARP-1, PARP-2, PARP-3, PARP-4, and PARP-5 [2]. Structure and pharmacological activities of the rest of the isoform need to discover further.
The structure of PARP mainly consists of four domains of interest.
A DNA-binding domain; A caspases cleaved domain; An automodification domain; A catalytic domain.
The DNA-binding domain further consists of two zinc finger motifs (Figure 1). When DNA damage takes place, DNA breaks bind to the DNA-binding domain. This binding leads to a conformational change. It has been shown that this binding occurs independently of the other domains. This is fundamental in a programmed cell death model based on caspase cleavage inhibition of PARP. After catalysis, the release of protein from DNA is done by auto-modification domain.
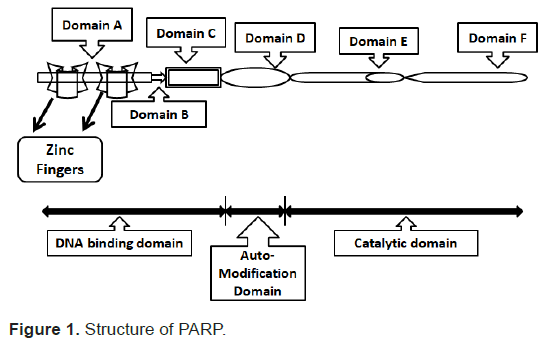
Figure 1: Structure of PARP.
PARP-1
PARP-1 is a most abundantly found nuclear protein which functions to sense the DNA damage. PARP-1 is a 116 kDa protein consisting of three main domains: the N-terminal DNA binding domain containing two zinc fingers, the automodification domain, and the C-terminal catalytic domain [3]. DNA breaks when bind to the DNA binding domain of the PARP; activate the PARP and causes dissociation of NAD into nicotinamide and ADP ribose. The activated PARP further polymerizes the ADP-ribose into nuclear acceptor proteins which include histones and transcription factors.
Moderate activation of PARP may reduce NAD content in the cell and can cause cell dysfunction. To prevent this dysfunctional stage and to maintain the normal physiology of the cells, pharmacological inhibition of PARP is required. Overactivation of PARP-1 causes apoptosis in response to damage conditions. It is necessary to prevent depletion of NAD for balanced apoptotic processes by inactivation of PARP-1.
Functions: PARP1 plays vital roles in various physiological and pathophysiological pathways including DNA damage signaling and cell death pathways. Due to which it is required for the maintenance of cell proliferation, differentiation, neuronal function, aging, inflammation, and carcinogenesis. PARP1 plays the following major functions; a) PARP1 have the property to ribosylate histones and can regulate chromatin structure. It helps in chromatin modification and transcription under physiological and pathophysiological conditions. b) Increased oxidative stress due to any pathophysiological condition in the body, leads to over-activation of PARP1. Over-activation of PARP1 decreases the amount of NAD+/ATP in the cell and also leads to translocation of Apoptosis-Inducing Factor (AIF) to the nucleus causing cell dysfunction and ultimately programmed cell death [4].
PARP1/poly-ADP-ribosylation-mediated cell death: PARP-1 acts on mitochondria and depending on the extent of oxidative stress, DNA damage, and PARP-1 activation, different cell death pathways may be triggered. A mild-to-moderate level of oxidative stress and PARP-1 activation may initiate cell death through a process involving mitochondrial depolarization/membrane permeability transition (MPT), resulting in the release of cytochrome c and Apoptosis Inducing Factor (AIF) from the mitochondrial intermembrane space into the cytosol. The AIF released from mitochondria moves to the nucleus and induces DNA fragmentation, which is considered an irreversible step in cell death. In pathological conditions where oxidative stress drastically increased and excessive DNA damage occurs, overactivation of PARP-1 take place and switches the mode of cell death from apoptosis to necrosis (Figure 2). This can possibly occur because of PARP-1-mediated excessive PARylation of apoptosis machinery and other essential proteins, resulting in their degradation and cell death [5].
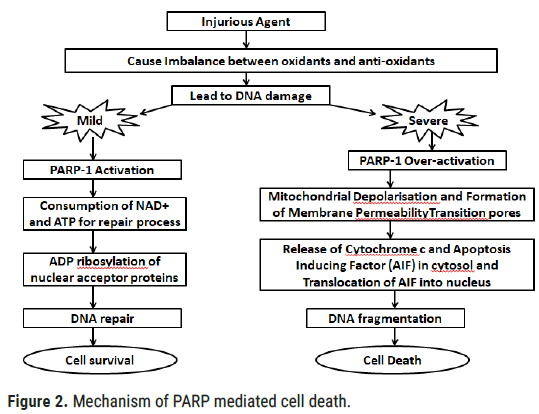
Figure 2: Mechanism of PARP mediated cell death.
PARP-2
PARP1, PARP2, and PARP3 are DNA dependent PARPs. They get stimulated by DNA damage. After PARP1, the second most studied subtype of PARP family is PARP-2. It was discovered after a long time after the invention of PARP-1. PARP-1 deficient cells were analyzed and found to exhibit 5% to 10% PARP activity which was because of PARP-2 [6]. Both PARP1 and PARP2 have overlapping cellular function and both are activated by DNA breaks. PARP-2 (like PARP-1) is a nuclear protein that binds to and is activated by DNAse-treated DNA. The DNA-binding domain of PARP-2 is slightly different from PARP-1 and could indicate distinct substrate specificities. The residual PARP activity was observed in PARP- 1 −/− cells after treatment with DNA-damaging agents. In research, PARP 2 was found to be a vital mediator of T-cell survival during thymopoiesis. This function can be due to the prevention of DNA damage-dependent apoptotic response [7]. Other than this, PARP-2 could function as an executioner of cell death pathways in focal and global cerebral ischemia.
PARP-3
The atomic mass of PARP-3 is 67 kiloDalton. It is the least studied and the the smallest PARP as compared to PARP-1 and PARP-2. It does not get activated upon DNA damage, further helps in DNA repair and promotes chromosomal rearrangements. PARP-3 is a major constituent of the centrosome, present in daughter centriole throughout the cell cycle. Magnification of PARP-3 interferes with G1 or S phase of cell cycle progression [8]. PARP-3 consists of a unique N-terminal domain of 39 amino acids. G4 DNA is a secondary structure when activated; it can form single-stranded guanine-rich DNA sequences. G4 DNA structures arise when four guanine bases interact through non-Watson–Crick base pairing to form planar G-tetrads. G4 DNA obstructs replication and repair. PARP3 regulates G quadruplex (G4 in response to DNA damage [9].
PARP-4
The atomic mass of PARP-4 is 193 kiloDalton. It lacks the N-terminal DNA binding domain. It is also known as vPARP because of the presence of large ribonucleoprotein complexes known as vaults, which are responsible for drug resistance and nucleocytoplasmic transport [10]. This isoform is present with mitotic spindle during cell division. Like poly (ADP-ribosyl) transferase, PARP-4 has a catalytic domain but lacks an N-terminal DNA binding domain due to which it does not a property to directly bind with DNA. The location and different morphology of these large cytoplasmic ribonucleoproteins emphasis their role in intracellular transport, particularly nucleocytoplasmic transport [11]. During multidrug-resistant human cell examination, the expression of these vault proteins was found to be increased, showing their role in intracellular detoxification [12]. Immunofluorescence and biochemical data show that PARP-4 is not exclusively associated with the vault particle but can also localize to the nucleolus, the nuclear spindle and to nuclear pores. The enzyme has also been found in association with mammalian telomerase but is dispensable for telomerase function and vault structure in vivo [13].
PARP-5 (Tankyrase)
PARP-5 is also known as tankyrase I. This protein was found to interact with telomeric-repeat binding factor-1 (TRF 1). Telomeric repeat binding factor is a negative regulator of telomere length [14]. Structurally, tankyrase I and Tankyrase II, both consists of stretches of consecutive histidine, proline and serine residues, followed by 24 ankyrin (ANK) repeats which is on N-terminus. The C-terminus is similar to PARP 1 catalytic region. DNA-Binding domain is absent in tankyrase, which suggests that its activity does not depend on the presence of DNA breaks. Tankyrase protein recruited to telomeres through binding to TRF1. This binding controls the telomere length [15]. Several knockdown studies suggest PARP5/tankyrase-1 as an isoform of PARP family which is required for mitosis in human cells. Removal of this gene can result in mitotic arrest and abnormal spindle structures [16]. In addition to its role in mitosis, PARP5/tankyrase-1 has also been suggested to require for the regulation of telomere length together with PARP6/tankyrase-2 by oligo ADP-ribosylation of the telomeric repeat binding factor TRF-1 [17].
PARP-6 (Tankyrase-2)
PARP 6 is also known as Tankyrase-2. It is found in the nucleus of human cells. Over-expression of it can lead to a release of endogenous TRF-1 from telomeres. Due to the catalytic activity of PARP, PARP 6 can function as a positive regulator of telomere length in human cells [18]. Both PARP 5 and PARP 6 regulate telomere extension in the presence of telomerase by poly-ADP-ribosylation in human cells [19]. PARP 6 plays an important role in normal growth, development, and metabolism. Growth retardation phenotype and reduced body fat can seem in PARP 6 knockout mice. PARP 6 is located in the cytosol and as a peripheral membrane protein associated with Golgi. It colocalizes with GLUT4 storage vesicles in the juxta-nuclear region of adipocytes. PARP 6 specifically binds to the GLUT4 vesicles and helps in regulating the activity of glucose transporter GLUT4 in adipocytes [20]. PARP6/tankyrase-2 has been shown to be a signaling target of Mitogen-Activated Protein Kinase (MAPK) and it gets phosphorylated upon insulin stimulation. Phosphorylation enhances the poly-ADP-ribose polymerase activity of tankyrase. In PARP6 knockout rodents, glucose uptake was found to be reduced without disrupting insulin-induced phosphorylation cascades [21].
PARG
There are 18 genes in total which are assumed to encode different PARP isoforms [22]. Out of which, there is an only single gene which encodes for the enzyme, responsible for catalyzing the hydrolysis of ADP-ribose polymers to free ADP ribose. It is known as Poly (Adp-Ribose) Glycohydrolase (PARG). It is the only enzyme known to catalyze the hydrolysis of ADP-ribose polymers to free ADP-ribose. It possesses both endoglycosidase and exoglycosisdic activity [23]. Its products are free Poly(ADP-Ribose) and monomeric ADP-ribose. Monomeric ADP-ribose is potent protein-glycating carbohydrate which has an ability to cause protein damage [24]. It helps in decreasing the risk of non-enzymatic protein glycation [25]. PARG prevents hypermodification of nuclear proteins with very long Poly-ADP-Ribose chains by modulating the complexity of poly-ADP-ribose on different acceptor proteins [26]. The degree of PARG activation is greater than PARP1 and PARP2, resulting in decreased PARP1 auto-modification and poly ADP-ribose accumulation. The genetic studies undoubtedly demonstrate that specific coordination of PARP and PARG activities is significant for cellular responses under normal physiological as well as cytotoxic stress conditions [27]. In another research, it is suggested that ADP-Ribose-Protein-Hydrolase-3 (ARH3) which is located in mitochondria, also possesses intrinsic PARG activity [28].
PARP plays an important role during changing the environment for cellular adaptations. Most of the diseases are because of over-activation of PARP-1 which leads to uncontrolled inflammatory mediator release, inflammation, adaptation, cell dysfunction, and further cell death. On view of this aspect, PARP inhibitors have gained interest of researchers so that they can be used as therapeutics to treat cell dysfunction, prevent cell organelle destruction, to treat oxidative stress-induced diseases and to prevent various other diseases associated with ageing such as neurodegenerative disorders, cardiovascular disorders, autoimmune and inflammatory disorders, stroke, and diabetes and their associated complications. The common cause of many of the above-mentioned diseases and conditions is an underlying inflammatory response. Thus, the therapeutic effects of PARP inhibitors in these diseases may share a common mechanism; namely, the inhibition of inflammatory pathways.
Conservative role in cardiovascular diseases
Cardiovascular disorders including circulatory shock, hypertension, atherosclerosis, arteriosclerosis, myocardial infarction, and ventricular hypertrophy occurs due to endothelial function. During endothelial dysfunction, PARP gets an over-activated lead to translocation of Apoptosis-Inducing Factor (AIF) from mitochondria into the nucleus. Chronic treatment with the PARP inhibitors PJ-34 and INO-1001 for 2 months in a rodent model has been demonstrated to improve endothelial and cardiac dysfunction associated with aging, showing the involvement of the nitro-oxidative stress–PARP pathway in the pathophysiology of cardiac and vascular aging. Furthermore, recent data suggest that activation of PARP importantly contributes to the development of endothelial dysfunction in various experimental models of diabetes and also in humans [29] and the pharmacological inhibition of PARP improves endothelium-dependent relaxation in these pathological conditions. PARP inhibition probably does not reduce initial damage but diminishes the ongoing formation of reactive compounds like NO* and subsequently peroxynitrite. PARP inhibitors block the translocation of AIF from the mitochondria into the nucleus. Thus, necrosis is reduced as well as overall cell death. Inflammation (neutrophil activation) is suppressed and the resulting morphological changes like hypertrophy are diminished. These all together lead to improved functionality of the heart and a reduction in infarct size [30].
Pharmacological inhibition or genetic inactivation of PARP helps in treating the myocardial necrosis in the acute and delayed stages of myocardial infarction and shows cardioprotective effects. These effects were seen in isolated heart preparations due to reduced catabolism of myocardial NAD+ as well as the preservation of the myocardial ATP stores. In another in vivo studies, beneficial effects were seen which were due to suppression of neutrophil infiltration in the absence of functional PARP. Free radical generation and oxidative stress continuously activate PARP in reperfused myocardium leading to DNA breakage. When DNA breakage persists for a longer period of time and remains unrepairable, it further causes PARP overactivation even in delayed myocardial reperfusion. The location of the most prominent PARP activation in the area of necrosis and peri-infarct zone in cardiac myocytes, which was highlighted by staining. Treatment with PARP-1 inhibitor (3-aminobenzamide) sufficiently suppresses PARP activation in the heart [31].
In a study, Spontaneously Hypertensive Rats (SHR) was used to check the role of PARP in hypertension. Animals were treated for 20 weeks with vehicle or the potent PARP inhibitor PJ34. In the vehicle-treated SHR rats, there was a significant loss of endothelial function when measured by the relaxant responsiveness of vascular rings to acetylcholine. SHR rats also developed severe hypertension and cardiac hypertrophy. Treatment with the PARP inhibitor did not influence high blood pressure and cardiac hypertrophy in SHR rats, but it improved Ach-induced, NO-mediated vascular relaxation. In addition to the beneficial effects of chronic treatment with the PARP inhibitor, 1-h in vitro incubation of aortic rings from SHR rats with PJ34 (3 μmol/l) was also able to improve the endothelial dysfunction [32].
Imbalance of PARP activity leading to carcinogenesis: In cancer chemotherapy, drug resistance in tumor cells is common because of the failure of apoptotic pathways and intracellular drug detoxification. Highly resistant tumor cells due to genetic alterations in caspases avoid apoptotic cell death pathways [33]. For apoptotic cell death pathway, high energy is required. But the cancer cells have low energy and become resistant to programmed cell death. That’s why; these tumor cell lines antitumor drugs can only cause cell death by the process of necrosis. The idea of PARP-1 activation in response to DNA breaks and further causing cell death helps to investigate the possibility of PARP inhibition on DNA binding anticancer therapies [34]. These investigations revealed that PARP-1 inactivation may comprise a suitable way to improve the activity of DNA-binding antitumor compounds because PARP-1 function is not essential for cell survival in the absence of extensive cell damage [35]. A recent study suggests that cells can be protected from necrosis by inhibition or inactivation of PARPs (e.g., PARP-1 and PARP-5b) so that the necrotic process is amenable to pharmacological intervention [36]. Inhibition of PARP-1 in cells which are exposed to DNA-damaging drugs will lead to decreased DNA repair and ultimately there will induction of apoptotic cell death by decreasing necrotic cell death and preventing the spillage of cell debris in the surrounding. It is interesting to note that PARP inhibitors might be more effective against tumor cells than against normal cells.
PARP and inflammatory diseases: The role of PARP activation and the protective effects of PARP inhibitors have also been demonstrated in various experimental models of inflammation, including acute inflammatory diseases (such as endotoxic shock), as well as chronic inflammation of the gut, joints, and various other organs. PARP inhibitors are found to be effective in the treatment of rheumatoid arthritis, ulcerative colitis, pulmonary inflammation and systemic inflammatory diseases (circulatory shock). The mechanism of PARP inhibitors for the treatment of inflammatory disorders is downregulation of pro-inflammatory mediators. The inhibition of PARP also reduces the formation of nitrotyrosine, an indicator of reactive nitrogen species in inflamed tissues. This finding was, at first, unexpected because PARP activation is perceived to occur downstream from the generation of oxidants and free radicals in various diseases. The mechanism is probably related to the fact that PARP inhibition reduces the infiltration of neutrophils into inflammatory sites. This reduces oxygen and nitrogen-centered free-radical production. The basis for the regulation by PARP of neutrophil infiltration might be related to the reduced expression of adhesion molecules and/or the preservation of endothelial integrity [37].
Incubation of type II pneumocytes with hydrogen peroxide leads to a decrease in phosphatidylcholine production. Inhibition of PARPs with 3AB or nicotinamide before exposure rescued energy levels as well as phosphatidylcholine synthesis [38]. Upon intratracheal administration of the DNA damaging agent bleomycin to mice, Genovese et al. could show that the two PARP inhibitors 3AB and 5 AIQ reduced the induced lung injury. Body weight loss and the mortality rate were diminished, as were edema formation and tissue injury. Nitrotyrosine levels as a marker of oxidative stress as well as myeloperoxidase activity (neutrophil infiltration) were significantly reduced [39]. A combination of smoke inhalation and Pseudomonas aeroginosa instillation into sheep lungs led to decreased functionality and increased tissue damage. Administration of the PARP inhibitor INO 1001 improved parameters like hemorrhage, congestion and inflammation scores and lowered oxidative stress [40]. Ischemia of isolated perfused rat lungs led to an increase of proinflammatory cytokines like TNF and IL1b, an increase in iNOS activity and subsequent elevated PAR formation with a drop in ATP levels [41]. Also, lung weight, permeability, and pulmonary artery pressure were increased. If the PARP inhibitor nicotinamide was given even 30 min after ischemia, the increase in all values was attenuated except for the pulmonary artery pressure, which was in fact slightly higher.
PARP activation contributes to various forms of reperfusion injury in the brain, heart, kidney, gut, liver and other organs. Selected ischemiareperfusion injury models in which PARP has a pathogenic role are diabetic nephropathy, skin, GI tract diseases, arthritis, etc [42]. Two organs in which PARP-mediated reperfusion injury has been intensively investigated are the brain and heart [43,44]. In both of these organs, treatment with pharmacological inhibitors of PARP and PARP1 deficiency reduces the infarct size and improve the functional outcome variables (such as neurological function and myocardial contractility). In the acute phase of the disease, the preservation of cellular energetic pools (and thereby the inhibition of parenchymal cell necrosis) is the mode of protection. Additionally, in the delayed stage of the disease, PARP inhibition prevents the activation of various inflammatory pathways. Direct evidence of PAR accumulation in reperfused organs has been provided by the immunohistochemical detection of PAR polymers [45]. In normal parenchyma, low levels of PAR accumulation are seen, whereas, in the ischaemic border zone (the area of risk in the heart or the penumbra in the brain), PARP activation can be seen. These cells might be in a state in which PARP is activated and the cells are dysfunctional but not yet necrotic. In the core of the infarct, necrotic cell death occurs. It has not yet been determined whether PAR or Poly(ADP-Ribosyl)ated proteins have extracellular signaling roles.
Involvement in metabolic disorders: Streptozocin is a compound that induces diabetes, causing a massive and selective killing of pancreatic β cells, through a mechanism of DNA alkylation and induction of the production of NO. This type of induction of disease is used as a model to study the disease in animals [46]. In response to the streptozotocin, β cells enter apoptosis, but recent studies have shown that the main route of the destruction of these cells for IDDM is necrosis. When cell death occurs by necrosis, the release of cytokines, NO, and ROS leads to inflammation and an increase in protein glycation, which results in an increased level of blood glucose. There is the possibility of using chemical inhibitors of PARP in the treatment of diabetes; this is exemplified by treatment with the pharmacological PARP inhibitor PJ-34 treatment. One week after the induction of diabetes, this drug ameliorated vascular PARP accumulation and restored normal vascular function, without altering systemic glucose levels, plasma glycated hemoglobin levels, or pancreatic insulin content [47]. Recent data indicate that pharmacological inhibition of PARP might suppress NF-680 κB activation and the expression of adhesion molecules both under constant high-glucose conditions in cultured endothelial cells in vitro. Another aspect of diabetes is the possibility of regeneration of the islets of Langerhans. It was demonstrated that PARP-1 is a component of the active transcription complex for the Reg gene and regulates its DNA binding by Poly(ADP-Ribosyl)ation [48]. The Reg protein is important for the proliferation of β cells and is induced by the concerted action of dexamethasone and IL-6. Thus, PARP-1 inhibition in combination with dexamethasone and IL-6 leads to increased transcription of Reg and accelerated the regeneration of pancreatic islets cells [49].
Poly (ADP-Ribosylation) helps in conversion of β-NAD+ into ADP-ribose, which binds to acceptor proteins and form polymers of different length and structure. This action is ensured by the mutual activity of Poly (ADP-Ribose) Polymerase (PARP) and Poly(Adp-Ribose) Glycohydrolase (PARG) enzymes, which are responsible for polymer synthesis and degradation, respectively. Poly (ADP-Ribosylation) also helps in sensing and repairing DNA damage generated by reactive oxygen species. But over activation of PARP causes NAD depletion, decrease ATP levels and further necrosis which are the leading cause of the inflammatory condition in many diseases. In this aspect, Inhibition of PARP enzyme could exert a protective role towards a number of pathological conditions [50].
PARP inhibitors
Benzamide: It is a well-known inhibitor of Poly(Adp-Ribose) Polymerase. It is the first generation PARP inhibitor. The Benzamide inhibit PARP by interfering with the binding of NAD to the enzyme's active site, but the benzamides also bind to DNA and thereby prevent the recognition of DNA breaks by PARP, inhibiting the activation of PARP [51]. Benzamide and its derivatives are more effective than nicotinamide, particularly 3-methoxy and 3-AB (IC50=22 mM for benzamide). They are less soluble in water due to their hydrophobic structure. Introduction of additional nitrogen ring improves the solubility of benzamide. Benzamide is 3-4 times more inhibitory potent against PARP-1 than against any other related enzyme [52].
Nicotinamide: Nicotinamide is an amide of vitamin B3. It is the precursor of coenzyme β-nicotinamide adenine dinucleotide (NAD+). NAD+ is considered to be necessary for cellular functions and metabolism. Nicotinamide is a weak PARP-1 inhibitor (inhibitory concentration for 50% reduction of PARP-1 activity, IC50=210 mM) and, moreover, it is not always an inhibitor but often an enhancer of ADP-ribosylation when administered to cells. At millimolar concentrations, nicotinamide interferes with NAD+ synthesis and becomes a substrate of NAD+ metabolic enzymes [53].
Dihydroisoquinolinones and isoquinolinones: The good results were obtained by using nicotinamide, benzamide, and 3-aminobenzamide as a PARP-1 inhibitor. This adds knowledge about the role of PARP-1 in various pathological conditions and key cellular processes. Further study led to the invention of some structurally improved compounds known as dihydroisoquinolinones. To form these, carboxamide group was attached with a lactam ring following the following of 3-Aminobenzamide. These improved structures avoid numerous drawbacks of first-generation inhibitors [55]. Dihydroisoquinolinones and isoquinolinones are found to be more potent as compared to aminobenzamide as PARP-1 inhibitor. For this reason, further compounds which were developed as PARP-1 inhibitor later followed the basic structure of dihydroisoquinolinones and isoquinolinones.
Benzimidazoles and indoles: To increase the potency of the inhibitor, some structural changes were done. Carboxamide group was held between the amide proton and nitrogen by an intramolecular hydrogen bond at cisorientation. Inhibitory activity of these compounds is due to two amide protons out of which one forms intramolecular hydrogen bond and other act as a hydrogen-bond donor by interacting with the enzyme. Simple benzimidazole-4-carboxamides results in more active PARP-1 inhibitor than the classical inhibitor 3-AB. Inhibitory activity of this compound was further increased by inserting phenyl substituent at position 2. Cocrystallisation of a 2-aryl-substituted benzimidazole-4-carboxamide with the catPARP domain helped in accommodating the large cavity of NAD+ binding site by its r-aryl ring. This does not allow the PARP to deplete NAD+ and further cause depletion of ATP and cell necrosis. Therefore, this compound named NU 1085 was adopted as a standard PARP-1 inhibitor because of its high potency, good water solubility and easy synthesis [56].
Isoindolinones: Isoindolinones are another type of PARP inhibitors. In these compounds, again carboxamide group is attached to the five-membered rings. This kind of inhibitors showed significant results in the treatment of ischemia-reperfusion injury and inflammation by acting against the reactive oxygen species induced cell death. To increase the potency of the compound, it is fused with adenosine that helps in recognition site for PARP-1 [56].
Phthalazinones and quinazolinones: Phthalazin-1(2H)-one is a less potent PARP-1 inhibitor with an IC50 of 12 mM. To increase the potency of this compound, methyl, benzyl, and ethyl groups were substituted at position 4 within the ring [56]. Other compounds were synthesized from phthalazinones through their combination with second nitrogen ring. These were known as Quinazolin-4-ones. Quinazolin-4-one and quinazoli- 2,4-dione have modest PARP-1 inhibition activity with IC50 values of 9.5 and 8 mM, respectively [57].
Phenanthridinones: Phenanthridinone was reported as a potent PARP- 1 inhibitor [58]. Since then, several substituted 6-(5H) phenanthridin-6- ones have been synthesized, showing stronger PARP-1 inhibitory effects than the unsubstituted phenanthridinone. Phenanthridine-5-one were used to develop Cyclopenta(imn)phenanthridine-5(4H)-one derivatives which showed increased inhibitory potency over the parental compound [39]. Another example of phenanthridinones has a property to protect brain cells from ischemia and hypoxia in vitro, named as N-(6-oxo-5,6- dihydro phenanthridine-2-yl)-2-(N,N-dimethylamino)acetamide (PJ 34). For this reason, it can be used in clinical purposes for treatment of various inflammatory disorders like transplanted organ function and allergic encephalomyelitis [59].
Zinc-fingers PARP-1 inhibitors: While studying the structure of PARP, we have come across two zinc fingers F1 and F2 which were present in the DNA binding domain. The detailed study suggested that Zinc F1 is only responsible for the activation of PARP-1 in the response of singlestrand DNA break whereas another zinc finger F2 does not promote PARP-1 activation at the time of DNA break. This study emphasized the researchers to develop inhibitors which can act on F1 zinc finger and inhibit the activation of PARP-1. The inhibitors were produced by oxidation of compounds that contain amino groups and lead to the production of C-nitroso compound which can only oxidize first zinc finger and further lead to inactivation of PARP-1. These compounds are being used to induce apoptosis in tumor cells and have been recently investigated for the treatment of cancer and AIDS [60].
Over the last decade, a multitude of studies have verified the role of PARP activation in a wide range of pathophysiological conditions. Furthermore, a series of animal experiments have proved that PARP-inhibition therapy represents an effective approach to treating a variety of diseases. The key to this remarkable effectiveness lies in the fact that PARP inhibition targets a relatively late event of oxidative cell injury. Therefore, the therapeutic window of intervention is quite wide, as indicated by the success of post-treatment regimens in some models. The wide variety of disease models in which PARP inhibition proved beneficial also indicates that PARP inhibitors block a common pathway(s) of tissue injury, such as NF-B activation or oxidative stress-induced cytotoxicity. Further work needs to establish the exact in vivo mechanism of action of PARP inhibitors. The information available to date supports the view that PARP over activation during inflammatory damage is a pivotal feature of tissue damage in various inflammatory-based pathologies and that the pharmacological inhibition of PARP may provide significant benefits in these conditions by salvaging affected tissues from necrosis and by reducing, as well as by downregulating the inflammatory responses. As with all new therapeutic areas, the usefulness of this target is unproven, but the potential effect of this class of agents is large.
Acknowledgement: This article was supported by Department of Pharmacology, School of Pharmaceutical Sciences, CT University, Ludhiana. We thank our mentor for assistance and comments which greatly improved the manuscript.
Author(s) contribution: Ms. Mandeep Kaur, wrote and designed the manuscript. Dr. Saurabh Sharma, supervised the drafting, critical revision and final version of manuscript to be published.
Conflict of interest: Authors have no conflict of interest.
Citation: Kaur M, et al. Insight on Poly (ADP-Ribose) Polymerase (PARP) as a Potential Pharmacological Target in Pathological Conditions. J Biol Today's World, 2021, 10(3), 001-006.
Received: 12-Feb-2021 Published: 05-Mar-2021, DOI: 10.35248/2322-3308.21.10.008
Copyright: © 2021 Kaur M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.